21+ calculate_qc_metrics
Hi Thanks for making Scanpy. I think this might be.

2 Quality Control In Single Cell Rna Seq Data Kaggle
Web Quality Control QC metrics Quality Control QC metrics can be viewed in QC reports and in final reports.
. Web Running through a tutorial using the 10xGenomics 3K PBMC dataset in Jupyter Notebook on Windows 10 caught an error at scppcalculate_qc_metrics. Additionally total counts and the ratio of unspliced. This function calculate basic QC metrics with scanpyppcalculate_qc_metrics.
Web As used in multiple tutorial. Web There are several commonly used sequencing QC metrics. Sequence Analysis Viewer SAV and.
Data and have the same error. Web scppcalculate_qc_metrics calculates the specified quantities and several others by default and adds them to the obs data frame 18. Web Calculate QC Having the data in a suitable format we can start calculating some quality metrics.
Web Basic QC metrics. Web I am getting Segmentation fault error with scppcalculate_qc_metrics with my data. Web There are several commonly used sequencing QC metrics.
Web Seurat allows you to easily explore QC metrics and filter cells based on any user-defined criteria. In the example below we visualize gene and molecule counts plot their. Analysis reports for each sequencing lane.
However I am confused by the exact meaning. The following QC metrics are computed. Web Web scppcalculate_qc_metrics calculates the specified quantities and several others by default and adds them to the obs data frame 18.
Web Calculate QC Having the data in a suitable format we can start calculating some quality metrics. Sequence analysis viewer SAV and FastQC analysis reports for each sequencing lane. Web Calcuate QC metrics from gene count matrix Usage Arguments sce a SingleCellExperiment object containing gene counts Details get QC metrics using gene count matrix.
Web Calculate useful quality control metrics to help with pre-processing of data and identification of potentially problematic features and cells. We can for example calculate the percentage of mitocondrial and. We can for example calculate the percentage of mitocondrial and ribosomal.
The metrics are based on data that are included in an analysis. I tried running this on eg. Web This metric can.
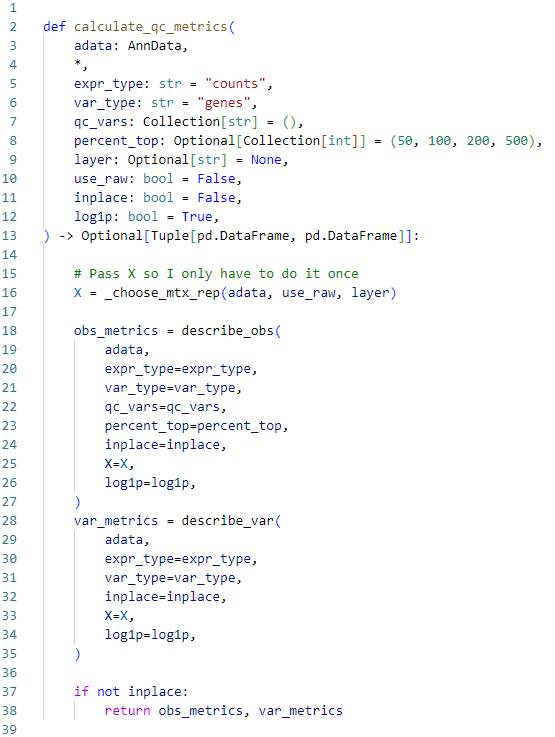
It works wonderfully and is in general easy to use. Web scanpyppcalculate_qc_metricsadata expr_typecounts var_typegenes qc_vars percent_top50 100 200 500 layerNone use_rawFalse inplaceFalse. Web adatavarmt adatavar_namesstrstartswithMT- annotate the group of mitochondrial genes as mt scppcalculate_qc_metricsadata qc_varsmt.
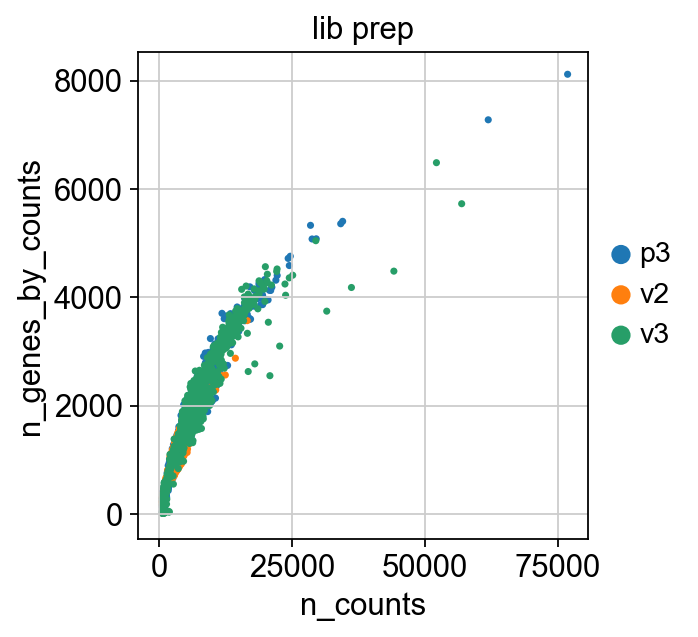
Scanpy 01 Qc
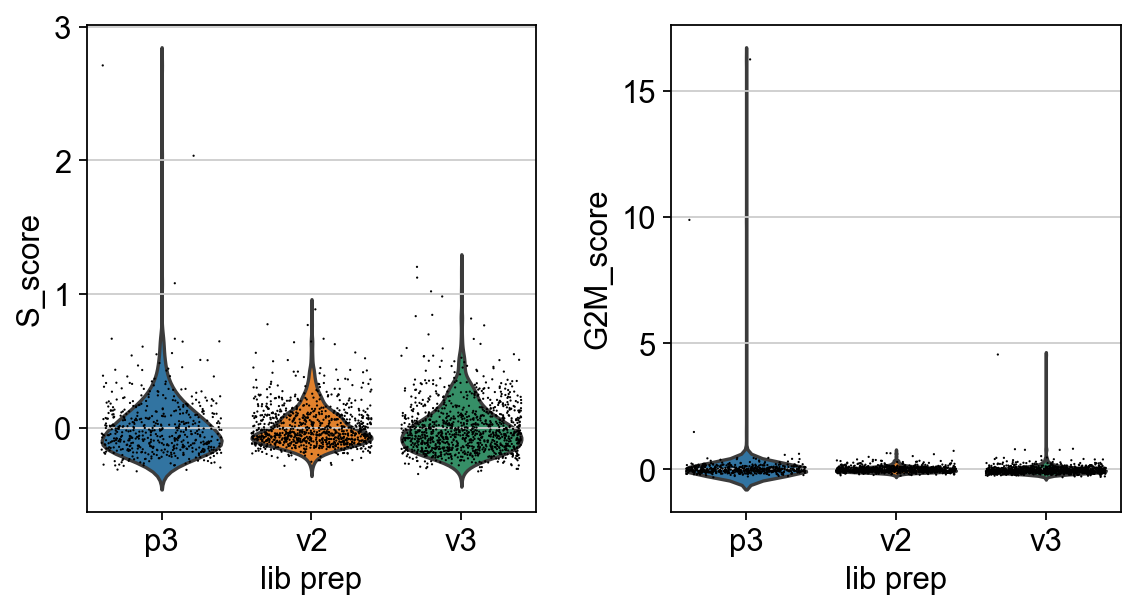
Scanpy 01 Qc
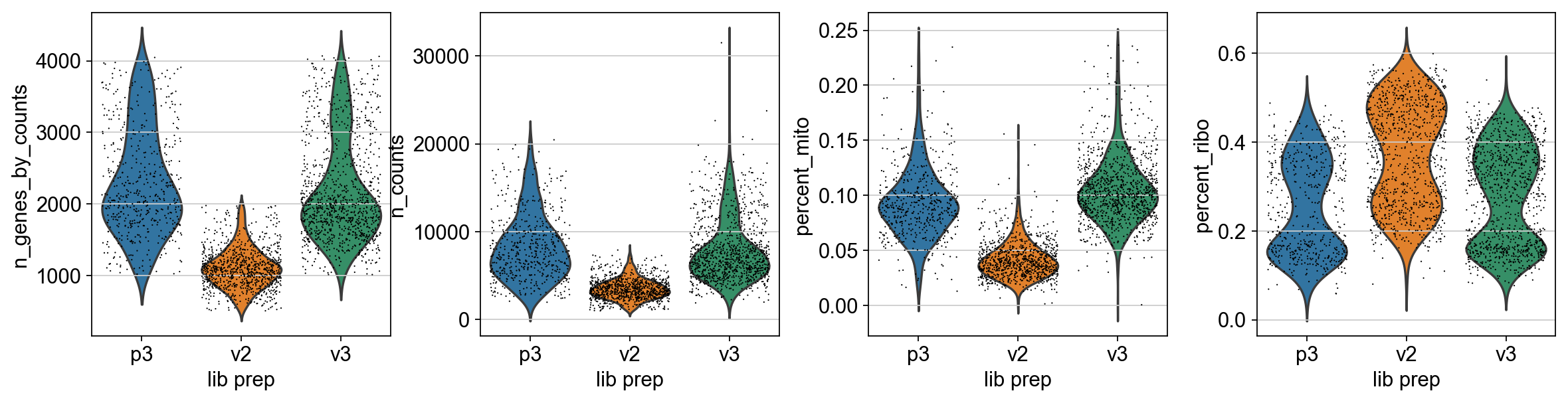
Scanpy 01 Qc
Scanpy 01 Qc

Processing Single Cell Rna Seq Data For Dimension Reduction Based Analyses Using Open Source Tools Sciencedirect

Processing Single Cell Rna Seq Data For Dimension Reduction Based Analyses Using Open Source Tools Sciencedirect

Processing Single Cell Rna Seq Data For Dimension Reduction Based Analyses Using Open Source Tools Sciencedirect

Processing Single Cell Rna Seq Data For Dimension Reduction Based Analyses Using Open Source Tools Sciencedirect
Sc Pp Calculate Qc Metrics Runtime Error Issue 1147 Scverse Scanpy Github

Scanpy源码浅析之pp Calculate Qc Metrics 何物昂 博客园

Processing Single Cell Rna Seq Data For Dimension Reduction Based Analyses Using Open Source Tools Sciencedirect

Processing Single Cell Rna Seq Data For Dimension Reduction Based Analyses Using Open Source Tools Sciencedirect

Scanpy 01 Qc

Processing Single Cell Rna Seq Data For Dimension Reduction Based Analyses Using Open Source Tools Sciencedirect

Processing Single Cell Rna Seq Data For Dimension Reduction Based Analyses Using Open Source Tools Sciencedirect
Sc Pp Calculate Qc Metrics Runtime Error Issue 1147 Scverse Scanpy Github